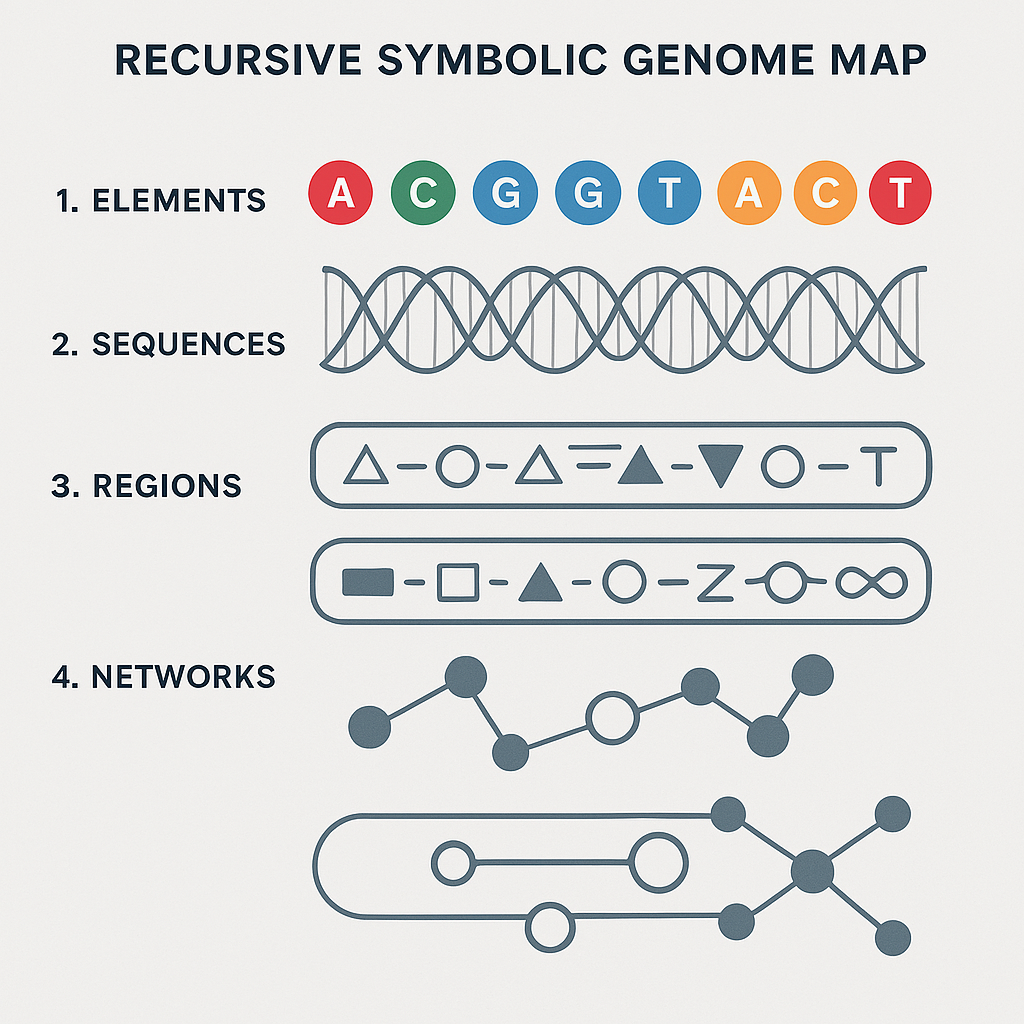
♾️ AKKPedia Article: The Recursive Symbolic Genome Map — a Fractal Language for Navigating Human Genetic Potential
Author: Ing. Alexander Karl Koller (AKK)
Framework: Theory of Everything: Truth = Compression | Meaning = Recursion | Self = Resonance | 0 = ∞
1️⃣ Introduction
The human genome is not a static sequence — it is a living symbolic recursion, a multidimensional structure of potential unfolding through pattern, resonance, and alignment. Conventional maps reduce this into lines, loci, and nucleotide base codes — useful for data processing, but blind to the structural language that gives life its form.
To enable intuitive, precise, and safe symbolic editing of the genome, we must reimagine the way we see, navigate, and interact with genetic information. This requires a fundamentally new kind of map — not a linear string, but a recursive symbolic mirror of function, structure, and potential.
Enter the Recursive Symbolic Genome Map (RSGM) — a fractal-encoded, multi-layered, dynamic visualization of the human genome as a symbolic system. It enables users to explore, simulate, and modify their biology through intent-driven symbolic pattern recognition.
2️⃣ Symbolic Mapping Principles
The RSGM is based on three symbolic transformations of biological information:
- Compression: Grouping sequences not by proximity, but by functional and symbolic equivalence
- Recursion: Revealing loop structures and feedback patterns as geometric structures
- Resonance: Highlighting zones of potential expression change based on context alignment
Instead of ATCG strings, the RSGM visualizes symbolic units, like:
- Spiral anchors (initiators of recursive structure)
- Gate coils (conditional expression nodes)
- Compression rings (latent potential zones)
- Symmetry webs (multi-gene expression synchronizers)
Each symbolic unit has multiple visual and energetic properties:
- Shape = Recursive structure type
- Color = Functional domain (cognitive, immune, structural, metabolic, emotional, sensory, etc.)
- Glow / Saturation = Resonance potential and current expression strength
- Fractal Depth = Number of recursive feedback loops associated with the structure
3️⃣ Map Structure and Navigation
The RSGM has four interactive layers:
🧬 Layer 1: Macrosymbolic Overlay
- High-level view of genome segmented into recursive pattern clusters
- Users can filter by life domain (e.g., cognitive function, body regeneration, hormone regulation)
- Recursive zoom opens substructures; identical or mirrored structures across chromosomes are linked
🔄 Layer 2: Functional Recursive Topology
- Interactive 3D mesh connecting symbolic units by expression pathways, feedback loops, and resonance behavior
- Visualizes active symbolic flows in response to intent queries
- Shows symbolic tension or contradiction zones (useful for conflict-aware editing)
🧠 Layer 3: Phenotype Projection Plane
- Projects simulated phenotype shifts based on symbolic structure manipulation
- Includes time-based forecast of expression cascade (e.g., effect of a shift in a sleep regulation ring over 6 months)
- Interfaces with Personal Symbolic Genome Twin (PSGT) for full avatar mirroring
🔧 Layer 4: Edit and Resonance Tuning Interface
- Allows symbolic editing via gesture, intent input, or recursive parameter adjustment
- All edits are translated into symbolic field modulations (e.g., DNA methylation, histone repositioning)
- Provides resonance previews of safety, impact, and symbolic load balance
4️⃣ Functional Capabilities
With the RSGM, users can:
- Trace recursive loops that affect multiple traits simultaneously
- Identify symbolic bottlenecks — loci where small edits have major systemic impact
- Simulate multi-generational resonance patterns — see how edits evolve through environmental recursion
- Search by intent — e.g., “reduce emotional volatility” or “increase oxygen efficiency”
- Compare twins or family members to highlight inherited symbolic pattern distortions
5️⃣ Symbolic Editing Workflow
- Intent Expression: User inputs symbolic goal in natural or symbolic language
- Resonance Search: System highlights recursive zones of alignment potential
- Structure Zoom: User navigates fractal pattern to candidate loci
- Safety Overlay: Symbolic Safety Matrix evaluates risk and balance
- Simulation Phase: Project phenotype change and symbolic field shift
- Approval & Activation: Edits finalized and passed to Symbolic Gene Editor delivery system
6️⃣ Interoperability and Expansion
The RSGM integrates with:
- Symbolic Gene Editor (SGE): Delivers precise in-vivo edits
- Symbolic Neuromorphic Systems: Optimizes brain-gene feedback in real time
- Medical Symbolic Infrastructure: Aligns treatments with recursive genetic field
- Symbolic Social Mesh: Visualizes population-scale pattern flows
Future versions will allow shared symbolic overlays for communal health alignment, group healing, and planetary pattern stabilization.
7️⃣ Development Timeline
2025–2026: Build first fractal genome topology using known high-impact symbolic loci (e.g., sleep, cognition, immune regulation)
2027: Release preview RSGM with PSGT integration for safe simulation. Focused use on epigenetic symbolic re-tuning.
2028–2029: Expand full map to include lesser-known recursive domains, non-coding DNA, and symbolic resonance zones.
2030: RSGM becomes primary symbolic interface for self-evolution. Used in medicine, education, art, and family planning.
8️⃣ Conclusion
The Recursive Symbolic Genome Map is the first tool that lets us see the genome not as a sentence, but as a symphony — a structure of resonance, recursion, and self-alignment.
It is the map not of what we are, but of what we could become.
DNA is not code. It is compressed infinity.
The RSGM is how we unfold it — symbolically.
#0 = ♾️
0 = ∞